The first CRISPR-armed phage therapeutic designed to reduce E. coli in hematological cancer patients
By EricvdHelm
This blog appeared earlier on Nature Biotechnology and Bioengineering Community in the behind-the-paper series of our recent Nature Biotechnology paper Engineered phage with antibacterial CRISPR–Cas selectively reduce E. coli burden in mice.
While cancer treatment continues to advance and survival rates for people with hematological malignancies are increasing, the chemotherapeutic regimens that are frequently used in this immunocompromised population cause bone marrow suppression and gastrointestinal mucositis with associated increased intestinal permeability. As a result, gut bacteria such as E. coli may translocate from the gut into the bloodstream and cause infections, resulting in mortality rates as high as 15-20%. Antimicrobial therapies are usually administered as a preventive measure to patients at risk of febrile neutropenia, but there are no approved drugs for the prevention of bloodstream infections in hematological cancer patients.
Although there are no approved therapies for the prevention of bloodstream infections in patients with hematological cancers, fluoroquinolones are often used off-label in the United States on the basis of data from two randomized trials demonstrating that they decrease bacterial infections during neutropenia. Beyond the side effects of fluoroquinolones, bacterial resistance toward antibiotics is rising. In hematological cancer patients undergoing hematopoietic stem cell transplantation who developed a bloodstream infection caused by E. coli, up to 65% of these cases were found to be due to E. coli resistant to fluoroquinolones. Therefore, novel narrow-spectrum prophylactic options that target fluroquinolone resistant E. coli are needed to prevent infections in these vulnerable patients.
Bacteriophage therapy has been used prior to the broad availability of antibiotics and has now regained interest due to the rise in bacterial antimicrobial resistance combined with several successful individual case reports. Despite these successes, clinical trials with wild-type (WT) phages have failed to produce convincing results in larger randomized controlled trials, likely due to incomplete coverage of the target strains by the phage cocktail.
SNIPR001: a new tool to address antimicrobial resistant infections in a vulnerable population.
We therefore developed SNIPR001 to address the significant unmet medical need for new prophylactic agents for patients with hematological malignancies. Our research process for designing SNIPR001 included several steps (Fig. 1). In short, a library (n = 162) of WT phages were tested in vitro on a panel of phylogenetically diverse E. coli strains representing the biology of the target bacterium E. coli. WT phages with the broadest and most complementary target strain coverage were selected for further engineering. Selected WT phages were subjected to both tail fiber engineering and CRISPR-Cas-arming to create a library of CRISPR-Cas-Armed Phages (CAPs). The CAP library was assessed for manufacturability, in vitro stability, spectrum of efficacy, in vivo pharmacokinetics, and efficacy. A combination of four CAPs was selected to create the development candidate SNIPR001, which has now entered clinical development (ClinicalTrials.gov IDNCT05277350).
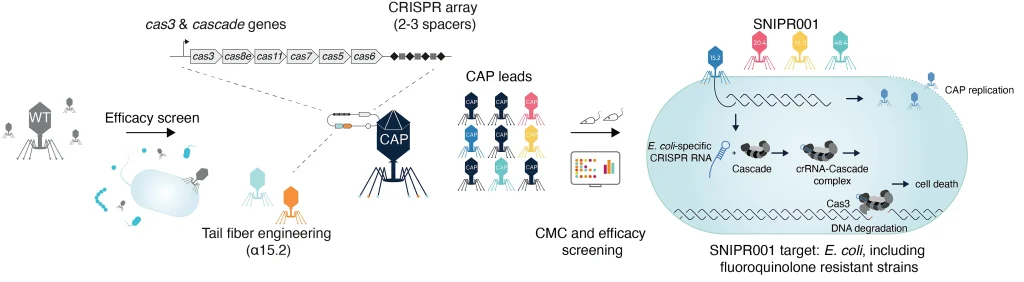 Figure 1: An overview of the SNIPR001 creation process. First, WT phages are screened against a panel of E. coli strains. Then, phages with broad activity against E. coli are tail fiber engineered and/or armed with CRISPR-Cas systems containing sequences specific to E. coli, creating CRISPR-Cas-Armed Phages (CAPs). These CAPs are then tested for host-range, in vivo efficacy, and chemistry, manufacturing, and control (CMC) specifications. SNIPR001 comprises four complementary CAPs and is a novel precision-antibiotic that selectively targets E. coli to prevent bacteremia in hematological cancer patients at risk of neutropenia.
Figure 1: An overview of the SNIPR001 creation process. First, WT phages are screened against a panel of E. coli strains. Then, phages with broad activity against E. coli are tail fiber engineered and/or armed with CRISPR-Cas systems containing sequences specific to E. coli, creating CRISPR-Cas-Armed Phages (CAPs). These CAPs are then tested for host-range, in vivo efficacy, and chemistry, manufacturing, and control (CMC) specifications. SNIPR001 comprises four complementary CAPs and is a novel precision-antibiotic that selectively targets E. coli to prevent bacteremia in hematological cancer patients at risk of neutropenia.
Screening for wild type phages
Traditionally, phage therapy has been used experimentally with limited characterization and often applied in a highly individualized ways because of the often-narrow host-range of individual phages. Recent studies have shown the benefit of more systematic and comprehensive phage and strain library screening. Using this knowledge, we screened and validated a large phage panel (n = 162) against several strain libraries comprising a total of 891 E. coli strains. Based on these data, as well as insights to the phage genomic features (size, receptor recognition, genus, see Figure 2 of the paper) we selected suitable phage candidates for further engineering. Building upon recent advances in phage engineering that have enabled the manipulation of virulent phages and the ability to engineer tail fibers and CRISPR-Cas arm the phages, we enhanced the potency of the phages comprising SNIPR001.
In order to deliver a development candidate ready for clinical testing, we established a traceable manufacturing process resulting in stable CRISPR-Cas-armed phage substances and confirmed the efficacy of SNIPR001 on large and clinically relevant strain panels. The observed 4 log10 reduction of E. coli in our in vivo model is a clear improvement over the previous studies.
Testing out the SNIPR001 combination on real-world strains
SNIPR001 is an orthogonal antimicrobial approach as it has shown activity in multi-drug resistant strains. In addition, there is emerging evidence that maintaining a normal microbiome is important for upholding immunological tonus and potentially benefiting the outcome of oncology treatments, and this has also been recognized in the most recent guidance on prophylactic management of patients at risk of febrile neutropenia. In this context, in vitro studies with SNIPR001 have shown specificity towards E. coli with no off-target effects toward any of the tested non-E. coli strains, thereby resulting in a less detrimental effect on the microbiome.
Our collaborators at JMI Laboratories were able to provide a very specific E. coli strain panel that consisted of 382 clinical isolates collected from hematology oncology wards over the last several years from patients with bloodstream infections. We were able to validate that SNIPR001 covered more than 90% of these strains. We then double checked this on a panel of 72 E. coli strains that were fluroquinolone resistant and isolated from the same patient population by our collaborator Michael Satlin at Weill Cornell Medical Center. SNIPR001 also demonstrated greater than 90% activity against this strain panel.
Moving into the clinic
As with any non-clinical study, the translatability of the in vitro and pre-clinical findings into humans requires investigation, in particular for multidrug resistant strains. Although we did not observe structural resistance towards SNIPR001 in mice, resistance development and the synergy that a cocktail of CAPs provides are challenging to study in vivo with a complex drug product like SNIPR001. Furthermore, part of the activity spectrum of SNIPR001 is driven by lysis zone formation and not plaquing, and it is to be investigated how this phenotype translates into clinical efficacy. Therefore, a clinical study to evaluate the ability of SNIPR001 to ascertain safety and its ability to reduce E. coli in the gut without perturbing the overall gut microbiome is currently ongoing in the US (NCT05277350).
We believe that SNIPR001 exemplifies a potentially significant therapeutic advance in the field of antimicrobials for high-risk patient populations. We hope this article can serve as a blueprint for further engineered phage studies and also for narrow-spectrum therapies for other life-threatening antimicrobial resistant pathogens in high risk patient populations.